Partners involved: CNR-IFC, CNR-INO, CNR-ITB, CNR-NANO, INFN, UNIPI
Coordinators: Valentina TOZZINI (CNR-NANO), Leonida A. GIZZI (CNR-INO), Maria Grazia ANDREASSI (CNR-IFC)
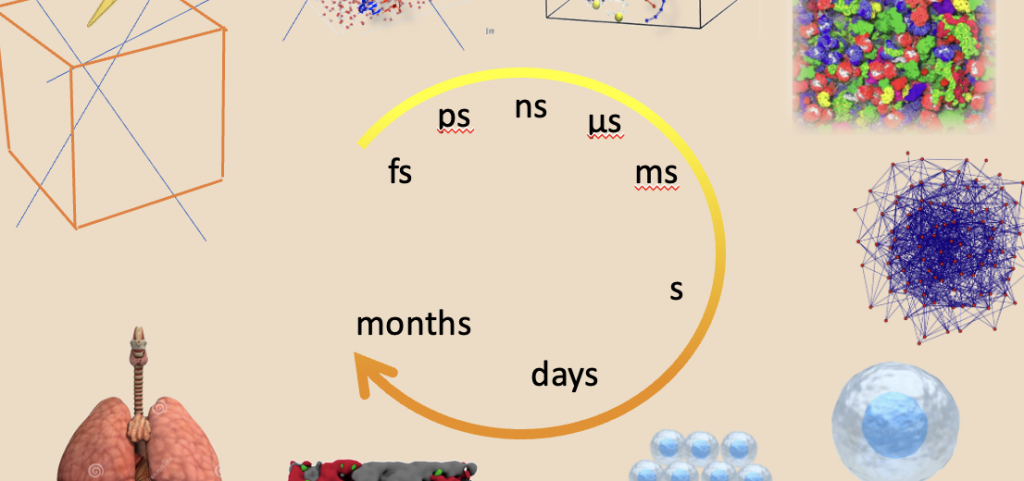
Outline: The underlying cellular mechanism of the differential effects of UHDR in cancer vs normal tissues are not fully understood. This issue will be addressed with a multi-scale and multi-methodological computer simulation and modelling approach. This in silico analysis will constitute a conceptual and quantitative bridge between the sub-project 1 and 3, with the general aim of clarifying the connections between the physical parameters of the radiation and the radiobiological effects in vitro. Currently, modelling approaches explore the molecular aspects by combining Monte Carlo (MC) simulations with Molecular dynamics (MD) atomistic simulations. This allows following the formation of the reactive species (ROS) and their effects over the ns scale. However, any differential effect either due to high dose or to the subsequent biomolecular cascade needs longer simulations (μs to ms) and sizes up to the (sub)cellular level. Therefore, atomistic simulations will be combined with low resolution models (coarse grained, CG) of the biomolecules embedded in a cytoplasm model accounting for the different compositions of the cells. The multi-scale method combined with experimental data from different sources should give a complete, yet detailed comprehension of the mechanisms of the UHDR differential effects. In turn, it should give indications for the optimization of the radiation beam characteristics (sub-projects 4 and 5), and for the radiotracers and radiopharmaceutical optimization (sub-projects 7 and 8). The models optimization and building on one side and the extraction of quantitative relationships useful for the source optimization and therapy planning on the other, require the elaboration of a large amount of statistical data from experiments and numerical simulations. Similar tasks are present also in other sub-projects. Therefore, sub-project 2 will be also devoted to the design and organization of a flexible database for data sharing and use. This task is of paramount importance to satisfy the main requirements of the PNRR calls and Horizon Europe program.
This subproject will also tackle in vitro validation of the mechanistic model based on the micronucleus assay, a standardized cytogenetic technique recommended by International Atomic Energy Agency (IAEA), that provides a quantitative measurement of radiation induced-chromosome damage (whole chromosomes or chromosome fragments) expressed as small nucleus at the end of mitosis in blood lymphocytes and on the cell survival in tumour cells and will define specifications for in vivo validation studies using VHEE beams in close collaboration with subproject 1.
Milestones of WP 1.2:
| M 1.2.1 – Design of the in silico multi-scale procedure and preliminary setup (software selection, materials acquisition) |
| M 1.2.2 – In vitro model validation for UHDR irradiation with LL VHEE beam and development of an experimental platform based on humanized 3D in vitro models |
| M 1.2.3 – Radiobiological effect simulations, and quantification of the radiation parameters/effect relationship by comparison with in vitro/in vivo. Database setup and implementation. |
